
Michael Watkins, PhD
Translational informatics, clinical interoperability, semantic engineering

Translational informatics, clinical interoperability, semantic engineering
2017–2021
2011–2017* Two year deferral (2012–2014), full-time volunteer mission for The Church of Jesus Christ of Latter-day Saints
2021–present
2023–present
2018–present
2018–2021
2017–2018
2016–2017
(a) Past |
|
| NLM/T15LM007124 | 07/13/18–07/01/21 |
| National Library of Medicine Training Program | $100,000 |
| Role: Trainee | |
| Department of the Interior | 01/01/20–06/30/22 |
| Pediatric Cancer Data Harmonization | $1,160,124 |
| Role: Other Personnel | |
(b) Current |
|
| St. Baldrick’s Foundation | 07/01/19–06/30/24 |
| Pediatric Cancer Data Commons | $2,700,000 |
| Role: Other Personnel | |
| NCI Leidos | 04/25/22–09/30/24 |
| Childhood Cancer Clinical Data Commons | $1,863,270 |
| Role: Other Personnel | |
| Gray Foundation | 05/01/22–06/30/24 |
| PREDICT—Monogenic Diabetes Data Commons | $89,500 |
| Role: Key Personnel | |
| Leukemia & Lymphoma Society | 11/01/23–10/31/27 |
| Dare to Dream | $2,997,218 |
| Role: Key Personnel | |
| MIB Agents | 07/01/23–06/30/24 |
| Data Commons Supported Precision Medicine | $100,000 |
| Role: Key Personnel | |
(c) Pending |
|
| NIH/U24 | 06/02/24–06/01/29 |
| RFA-CA-22-023 | $3,850,020 |
| Realizing a Unified Oncology Knowledge Graph | |
| Role: Key Personnel | |
Back-End Programming
Front-End Programming
Application Architecture
Clinical Interoperability
Ontologies and Terminologies
Clinical Decision Support
Semantic Engineering
Genomics
Analytics
| 2020 | Recipient, "John D. Morgan Award", University of Utah, Department of Biomedical Informatics |
External |
|
| 2017–2020 | Global Alliance for Genomic Health (GA4GH), Variant Representation Working Group |
| 2018–Present | Health Level Seven International, Clinical Genomics Working Group (Co-Chair) |
| 2018–2020 | Electronic Medical Records and Genomics (eMERGE) EHR Integration (EHRI) Work Group (Lead Investigator) |
| 2019–2021 | American Medical Informatics Association, Genomics and Translational Bioinformatics Working Group |
| 2022–Present | National Institutes of Health, National Heart, Lung, and Blood Institute, BioData Catalyst® Data Management Core, Data Standards Team |
University of Utah |
|
| 2018–2021 | Department of Biomedical Informatics, Curriculum Committee |
| 2018 | Department of Biomedical Informatics, Reed Gardner Teaching Excellence Award Committee |
| 2020 | Department of Biomedical Informatics, Industry Advisory Board Steering Committee |
| 2020 | Department of Biomedical Informatics, PhD Admissions Committee |
University of Chicago |
|
| 2022 | Comprehensive Cancer Center, EYES on Cancer, Research Team |
| 2022 | Data Science Institute, Summer Lab Program |
| 2021–Present | American Medical Informatics Association / Health Level Seven International, FHIR App Competition |
| Ad hoc review | Americal Medical Informatics Association Annual Symposium, BMC Med Informatics Dec Making |
| BMI 6030 | Graduate, “Foundations of Bioinformatics”, Course Developer, Co-Instructor, 2017–2020 |
| BMI 7902 | Graduate, “FHIR Practicum”, Course Developer, Co-Instructor, 2018–2021 |
| Workshop | Graduate, “Data Modeling Using FHIR”, Content Developer, Instructor, 2019–2020 |
| Workshop | Corporate, “HL7® FHIR® 101 for Developers”, Content Developer, Co-Instructor, 2019–2021 |
| Workshop | Corporate, “SMART on FHIR and CDS Hooks”, Content Developer, Instructor, 2020–2021 |
| Workshop | Corporate, “Introduction to CQL”, Content Developer, Instructor, 2020–2021 |
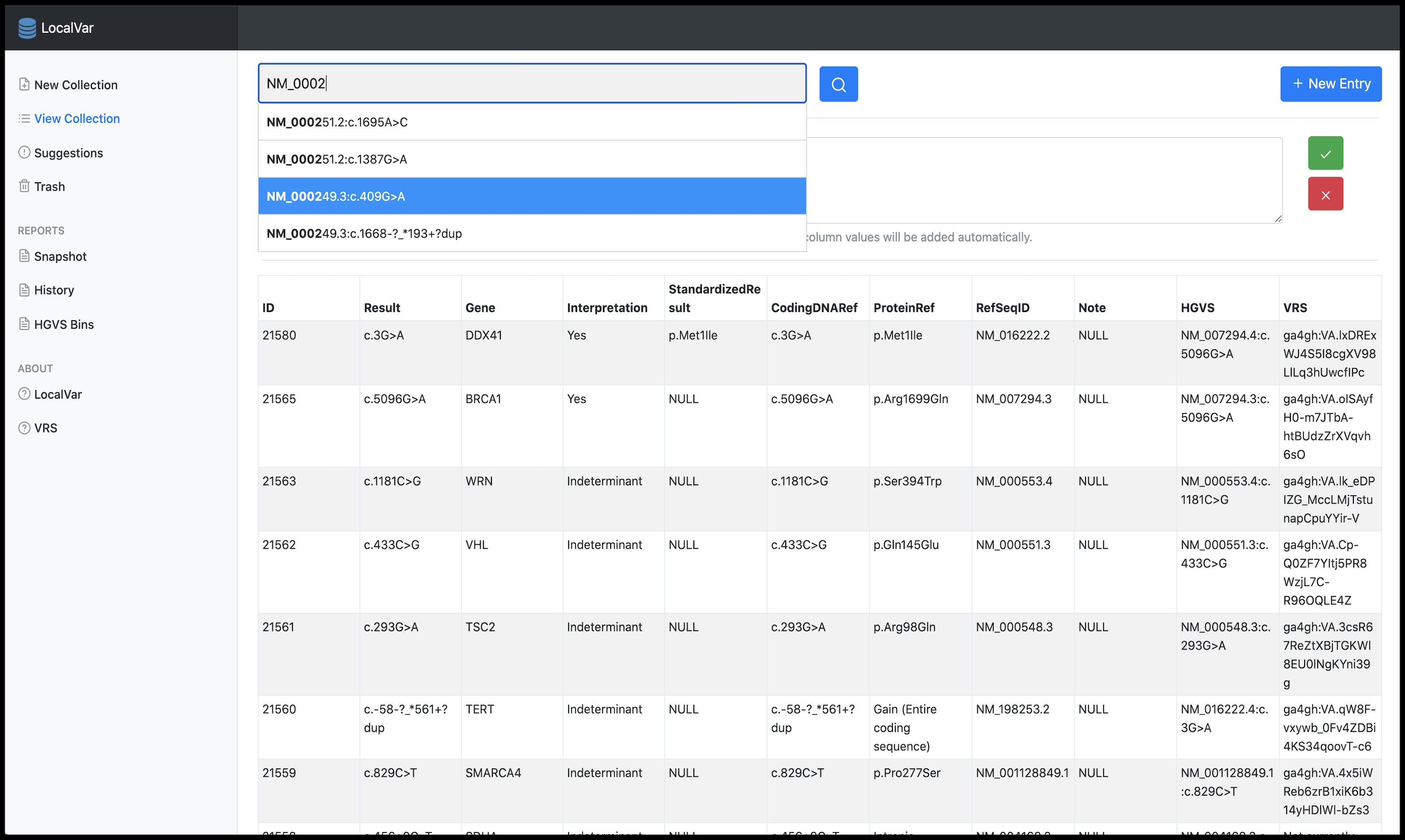
A local variant collection manger to asynchronously detect synonyms, HGVS expression changes, and variant interpretation changes from ClinVar.
View on Github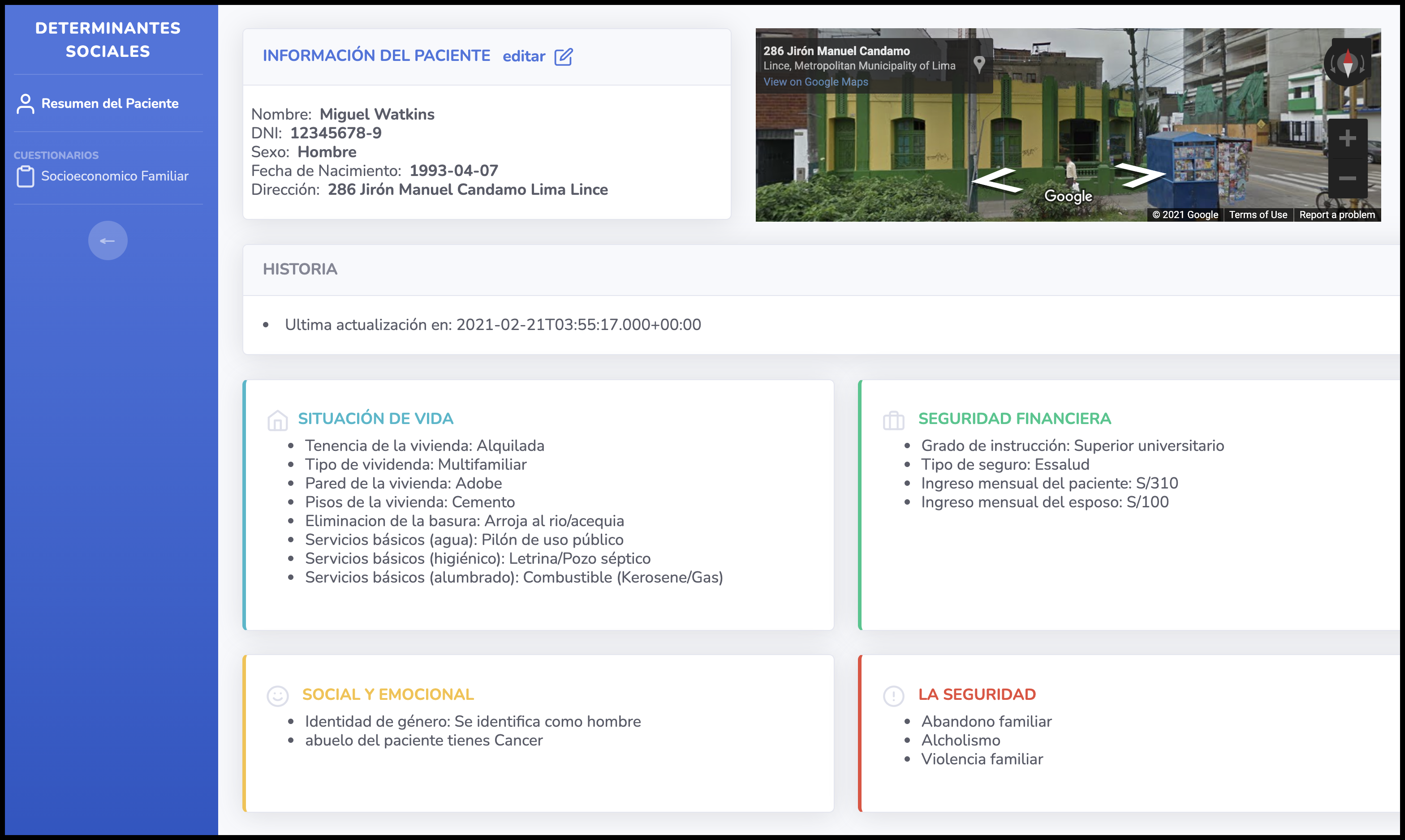
A FHIR-based app for collecting social determinantes of health via FHIR Questionnaire and displaying a categorical summary. Designed, built, and deployed for a partner clinic in Lima, Peru.
View on Github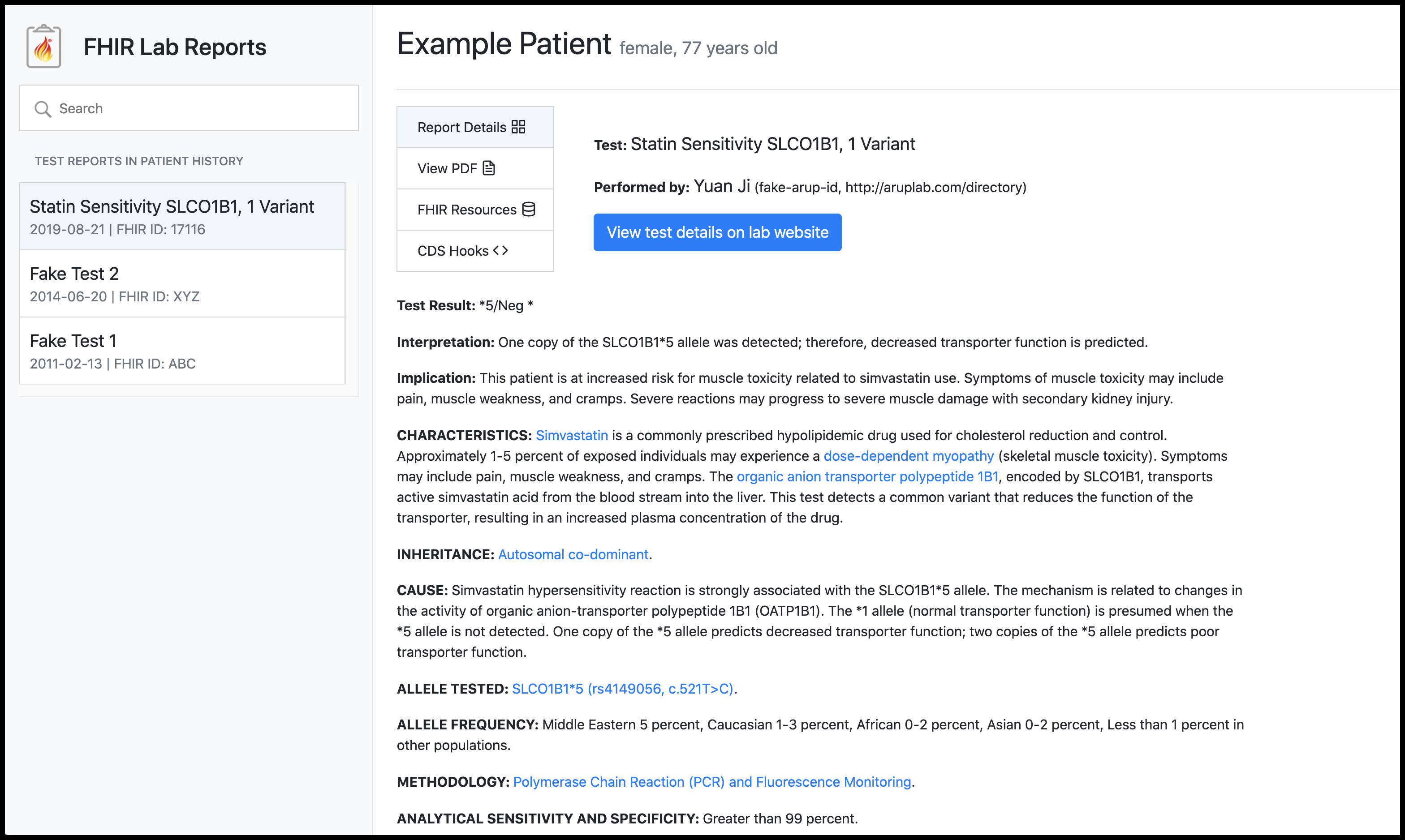
FHIR Lab Reports is a standards-based interoperable application that transforms medical laboratory test results into computable data points, transmits them to the ordering clinical system via SMART on FHIR, and makes them accessible to clinicians via CDS Hooks.
View on Github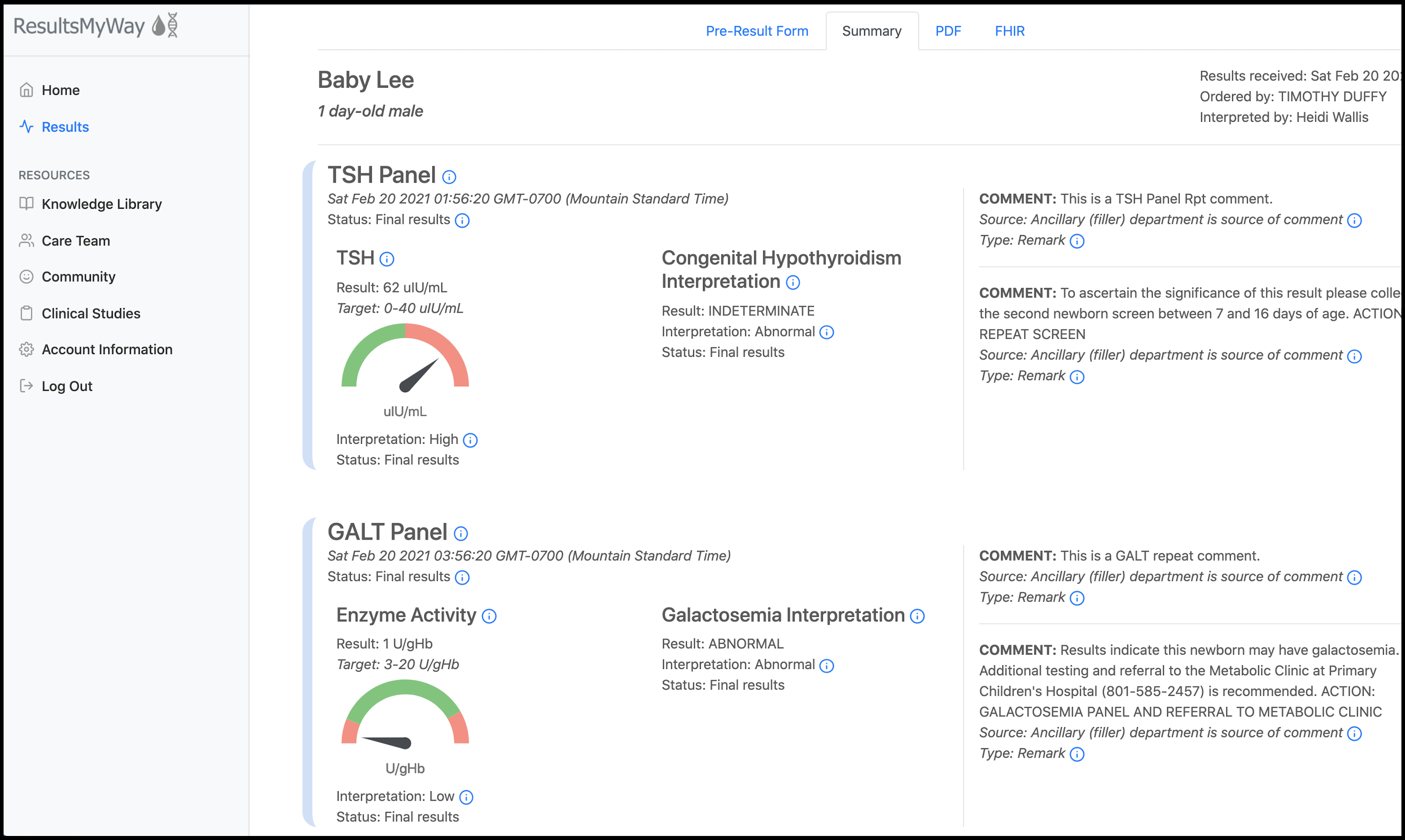
ResultsMyWay is a newborn screening (NBS) application that uses CQL to automate the search for maternal and infant health factors that may influence the interpretation of NBS results. It also translates those results to FHIR resources and acts as a hub for several types of informational resources about the rare disorder that a given infant may be diagnosed with.
View on Github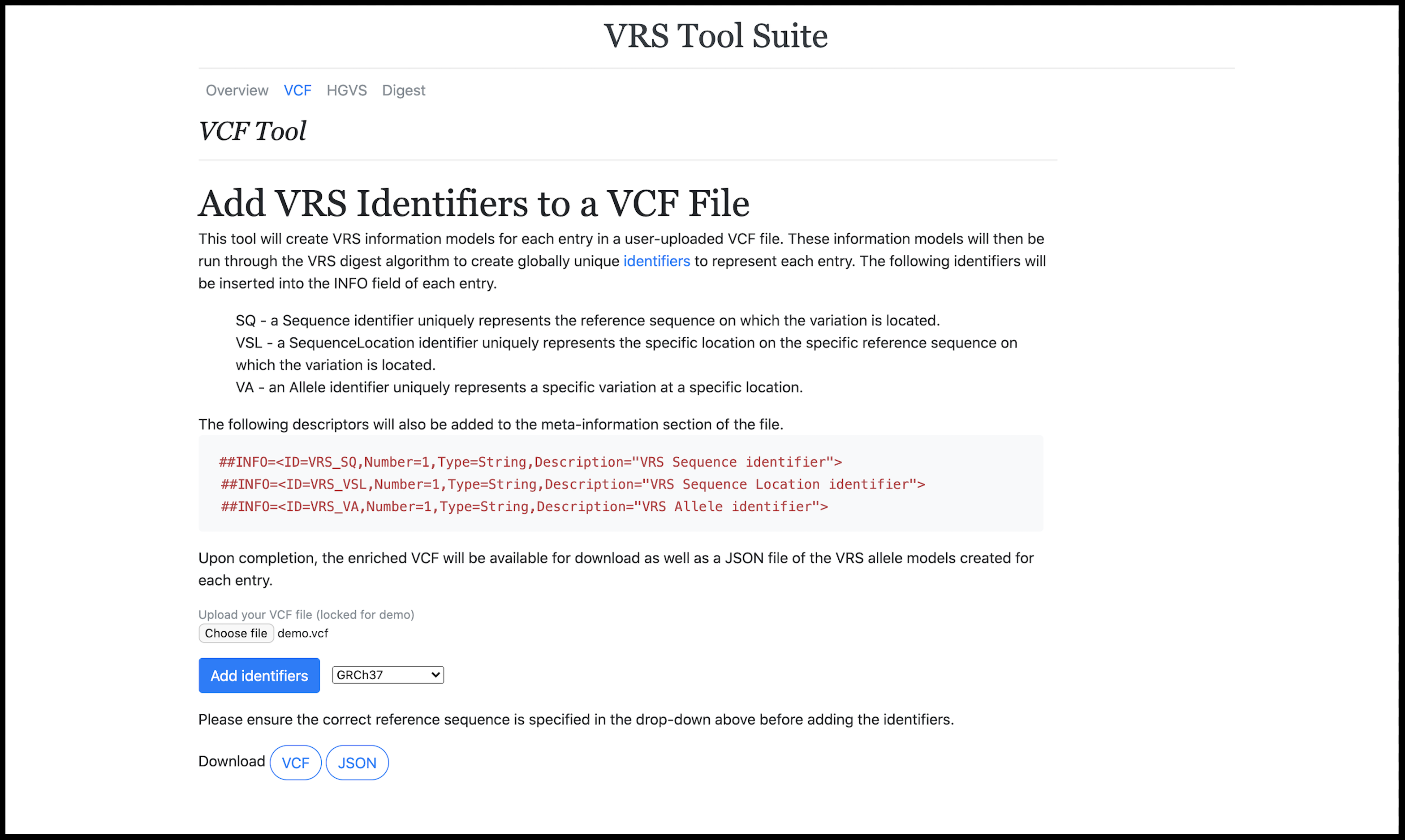
Three web tools that implement the GA4GH Variant Representation Specification (VRS). The first adds VRS identifiers to a VCF file, the second converts an HGVS expression to VRS identifiers, and the third allows input fields to create VRS identifiers for a custom variant.
View on Github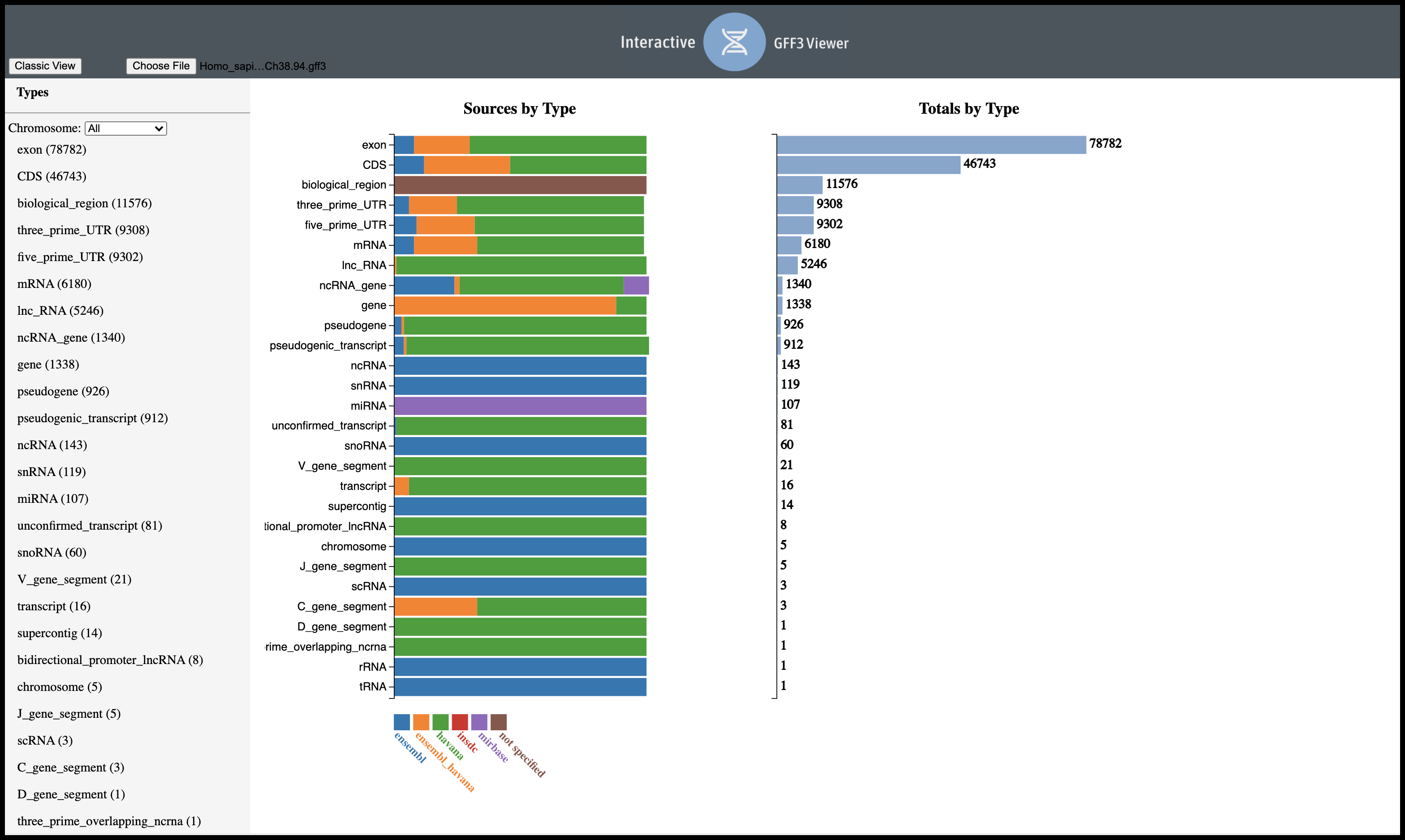
Facilitates the manual exploration of a gff3 file. Shows file-wide statistics, a search/filtering view for individual features, and a tree view to show parent/child relationships.
View on Github| 2018 | Invited speaker, “Precision medicine and its emerging app development landscape”, University of Utah Industry Advisory Board (IABTalks), Salt Lake City, UT |
| 2018 | Invited speaker, “FHIR queries and creating a FHIR-based medical calculator”, University of Utah School of Medicine DeCART Summer Program, Salt Lake City, UT |
| 2018 | Invited speaker, “Using genomic data in CDS via FHIR”, University of Utah Translational Research Informatics Special Interest Group, Salt Lake City, UT |
| 2018 | Invited speaker, “Biomedical Informatics and Graduate Studies”, Brigham Young University Life Sciences Career Exploration, Provo, UT |
| 2019 | Plenary talk, “Use of Infobuttons to find answers to clinicians’ questions in clinical genomics”, National Library of Medicine Informatics Training Conference, Indianapolis, IN |
| 2019 | Invited speaker, “FHIR, SMART, and Social Determinants of Health”, Pan American Health Organization (PAHO) and the Peruvian Ministry of Health, Lima, Peru |
| 2019 | Invited speaker, “Genomic Infobuttons—Matching info needs to electronic knowledge resources”, University of Utah Translational Research Informatics Special Interest Group, Salt Lake City, UT |
| 2019 | Invited speaker, “Creating a SMART on FHIR application and deploying it with CDS Hooks”, University of Utah IT Exchange (ITX) Group, Salt Lake City, UT |
| 2019 | Plenary talk, “Implementing the VMC specification to reduce ambiguity in genomic variant representation”, American Medical Informatics Association, Annual Symposium, Washington, DC |
| 2019 | Invited speaker, “How using the GA4GH VR-Spec can improve and promote data sharing”, University of Utah Molecular Data Warehouse (MDW) Group, Salt Lake City, UT |
| 2020 | Invited speaker, “GA4GH VR-spec overview and implementation in HCI”, Huntsman Cancer Institute, Population Sciences Variant Data Team, Salt Lake City, UT |
| 2020 | Plenary talk, “FHIR Lab Reports: using SMART on FHIR and CDS Hooks to increase the clinical utility of pharmacogenomic laboratory test results”, American Medical Informatics Association, Virtual Summit |
| 2020 | Plenary talk, “A SMART on FHIR and CDS-Hooks enabled approach to the exchange and review of genomic test results”, National Library of Medicine Informatics Training Conference, Indianapolis, ID |
| 2020 | Invited speaker, “IAB Student Lightning Talk”, University of Utah Industry Advisory Board (IABTalks), Salt Lake City, UT |
| 2021 | Plenary talk, “ResultsMyWay: combining Fast Healthcare Interoperability Resources (FHIR), Clinical Quality Language (CQL), and informational resources to create a newborn screening application”, American Medical Informatics Association, Virtual Summit |
| 2021 | Invited speaker, “FHIR Lab Reports: using SMART on FHIR and CDS Hooks to increase the clinical utility of pharmacogenomic laboratory test results”, University of Utah Morgan Award Acceptance, Salt Lake City, UT |
| 2022 | Invited speaker, “Genetic Analysis Elements for the PCDC Rhabdomyosarcoma Data Dictionary”, International Soft Tissue Sarcoma Consortium (INSTRuCT) Executive Committee Meeting, Virtual |
| 2022 | Invited speaker, “Pediatric Oncology and mCODE”, Childhood Cancer–Data Integration for Research, Education, Care, and Clinical Trials Partnership (CC-DIRECT), Virtual |
| 2022 | Plenary talk, “LocalVar: a local variant collection manager to asynchronously detect synonyms, HGVS expression changes, and variant interpretation changes from ClinVar”, American Medical Informatics Association, Annual Symposium, Washington, DC |
| 2023 | Invited speaker, “Some ‘Growing Pains’ of Health Data Interoperability”, Brigham Young University Life Sciences Graduate Seminary Series, Provo, UT |
| 2023 | Invited speaker, “AYA Represention in The Cancer Moonshot Initiative”, Teen Cancer America Moonshot Initiative Roundtable, Los Angeles, CA |
| 2023 | Invited speaker, “Genomic Alteration Modeling in Neuroblastoma”, International Neuroblastoma Risk Group (INRG) Annual Meeting, Amsterdam, Netherlands |
| 2024 | Invited speaker, “Developing Pediatric Cancer Data Standards”, National Cancer Institute Childhood Cancer Data Initiative (CCDI) Webinar Series, Virtual |
| 2024 | Invited speaker, “Pediatric Cancer Common Data Elements (CDEs)”, North American Association of Central Cancer Registries (NAACCR) Site-Specific Data Item (SSDI) Working Group Meeting, Virtual |
Watkins M, Fujimoto MS, Clement M. (2016) A Simplified Framework for Neural Networks [poster]. BIOT.
Watkins M, Rynearson S, Henrie A, Eilbeck K. (2019) Implementing the VMC specification to reduce ambiguity in genomic variant representation. AMIA Annu Symp Proc. 2019:1226-1235.
Watkins M, Eilbeck K. (2020) FHIR Lab Reports: using SMART on FHIR and CDS Hooks to increase the clinical utility of pharmacogenomic laboratory test results. AMIA Jt Summits Transl Sci Proc. 2020:683-692.
Watkins M, Viernes B, Nguyen V, Rojas Mezarina L, Silva Valencia J, Borbolla D. (2020) Translating social determinants of health into standardized clinical entities. Stud Health Technol Inform. 70:474-478. doi: 10.3233/SHTI200205.
Watkins M, Au A, Vuong T, Wallis H, Hart K, Rohrwasser A, Eilbeck K. (2021) ResultsMyWay: combining FHIR, Clinical Quality Language (CQL), and informational resources to create a newborn screening application. AMIA Annu Symp Proc.; 2021:615–623.
Watkins M, Au A, Eilbeck K, Ruiz-Schultz N, Rohrwasser A, Hart K, Williams M. (2022) A comprehansive newborn screening data interface for Utah providers, parents, and guardians [poster]. APHL.
Watkins M, Furner B, Li M, Cohen E, Carlson B, Leary S, Hettmer S, Kamihara J, Wyatt K, Volchenboum S. (2022) Harmonizing Genetic Data Element Modeling Across Cancer Trials [poster]. 54th Congress of the International Society of Paediatric Oncology (SIOP).
Watkins M, Kohlmann W, Berry T, Sama N, Koptiuch C, Rynearson S, Eilbeck K. (2023) LocalVar: a local variant collection manager to asynchronously detect synonyms, HGVS expression changes, and variant interpretation changes from ClinVar. AMIA Annu Symp Proc. ; 2023:1145-1152.
Carlson B, Watkins M, Li M, Furner B, Cohen E, Volchenboum S. (2023) Using A Standardized Nomenclature to Semantically Map Oncology-Related Concepts from Common Data Models to a Pediatric Cancer Data Model. AMIA Annu Symp Proc. ; 2023: 874–883.
Furner B, Cheng A, Desai A, Benedetti D, Friedman D, Wyatt K, Watkins M, Volchenboum S, Cohn S. (2024) Extracting Electronic Health Record Neuroblastoma Treatment Data with High Fidelity Using the REDCap Clinical Data Interoperability Services Module. JCO Clinical Cancer Informatics vol. 8 (2024): e2400009.
Wyatt K, Birz S, Castellino S, Henderson T, Lucas J, Pei Q, Zhou Y, Volchenboum S, Furner B, Watkins M, Kelly K, Flerlage J. (2024) Accelerating pediatric Hodgkin lymphoma research: the Hodgkin lymphoma data collaboration (NODAL). Journal of the National Cancer Institute. 2024;, djae013.
Watkins M, Christensen E, Eilbeck K (2024) Categorizing Genomic Concepts from Clinical and Research Terminologies Commonly Used in Cancer Reporting. AMIA Annu Symp Proc. 2024; Submitted Mar 2024.